
The header begins the file and provides metadata describing the body of the file.
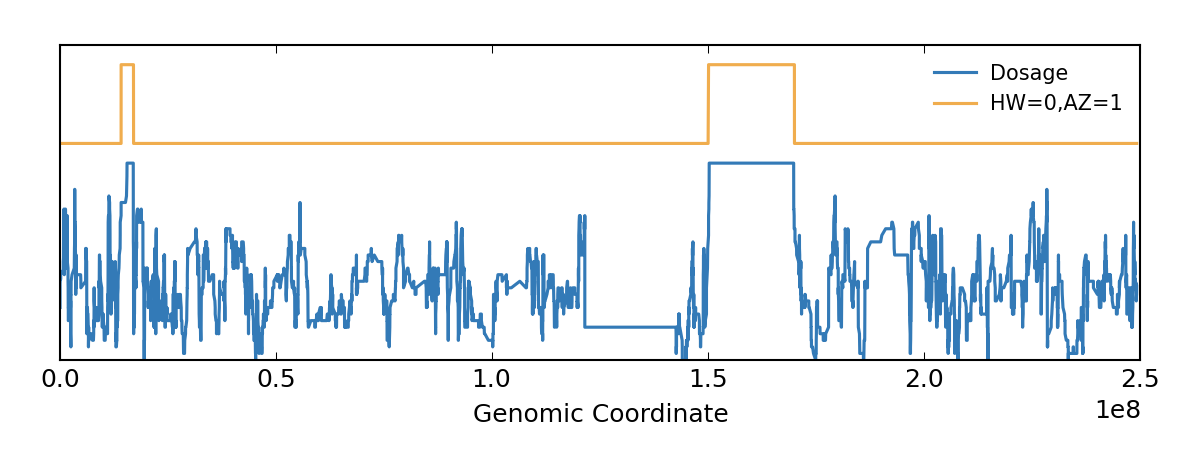
A set of tools is also available for editing and manipulating the files. There is also a Genomic VCF ( gVCF) extended format, which includes additional information about "blocks" that match the reference and their qualities. The standard is currently in version 4.3, although the 1000 Genomes Project has developed its own specification for structural variations such as duplications, which are not easily accommodated into the existing schema. By using the variant call format only the variations need to be stored along with a reference genome.
Existing formats for genetic data such as General feature format (GFF) stored all of the genetic data, much of which is redundant because it will be shared across the genomes.
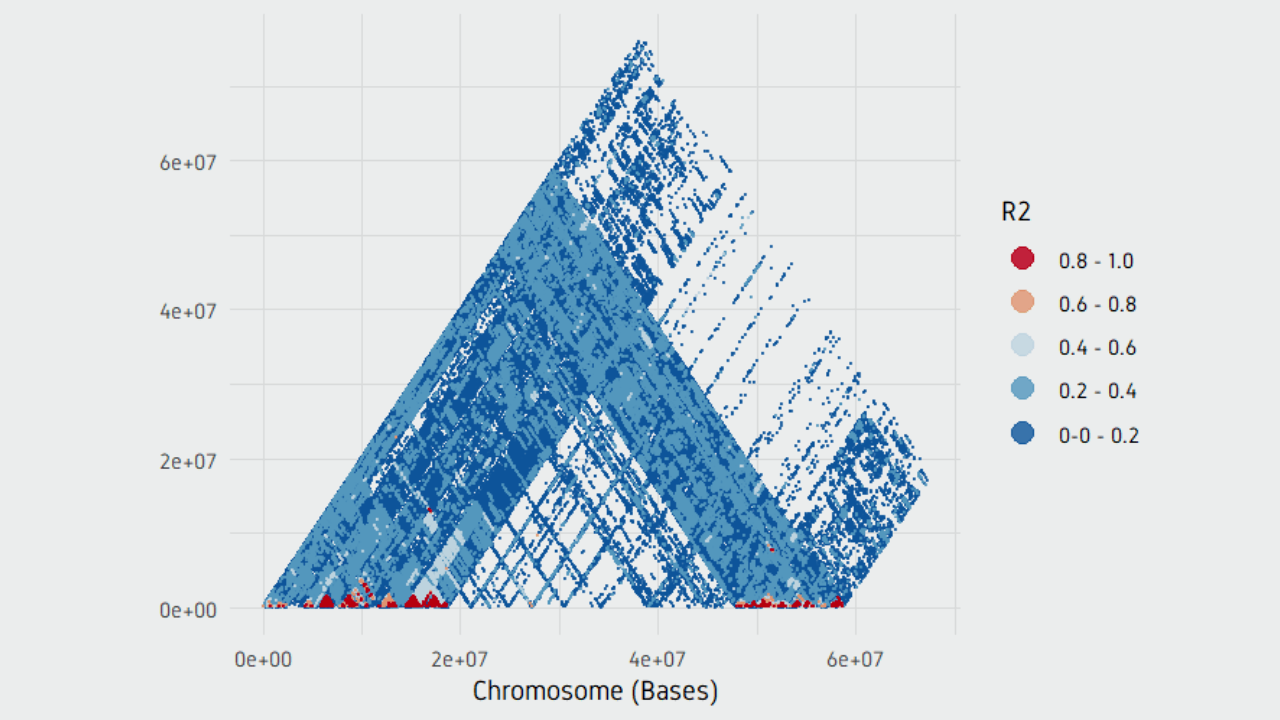
The format has been developed with the advent of large-scale genotyping and DNA sequencing projects, such as the 1000 Genomes Project. The Variant Call Format ( VCF) specifies the format of a text file used in bioinformatics for storing gene sequence variations. Demonstrating the difference between the binary BCF and VCF formats.


 0 kommentar(er)
0 kommentar(er)
